The Convergence Science Intercalated PhD Pathway
The Convergence Science Intercalated PhD Pathway
The iPhD programme is designed to equip clinical academics with the expertise to tackle complex challenges by integrating cancer research with engineering and physical sciences. Tailored for exceptional undergraduate students enrolled in the MBBS/BSc degree course, this programme presents the unique opportunity to pursue an intercalated PhD alongside their studies.
The programme is a pivotal component of the Cancer Research UK Clinical Academic Training Programme. This forward-thinking initiative is funded by the CRUK Convergence Science Centre, reflecting a strong commitment to advancing both medical and research frontiers.
This pathway offers a distinct advantage to PhD supervisors. The collaboration between clinical academics and research experts not only enriches the training environment but also brings diverse insights to research projects. The unique combination of medical insight and research rigor can foster innovative problem-solving and lead to a more comprehensive understanding of complex medical challenges. The supervisor benefits from a motivated student whose dual expertise can propel research projects forward, opening new avenues for exploration and breakthroughs.
In essence, the iPhD Pathway redefines academic training by seamlessly weaving together medical expertise and research acumen. By embracing this dynamic approach, students become not only skilled practitioners but also visionary thinkers driving advancements at the forefront of cancer research and its intricate connections with engineering and the physical sciences.
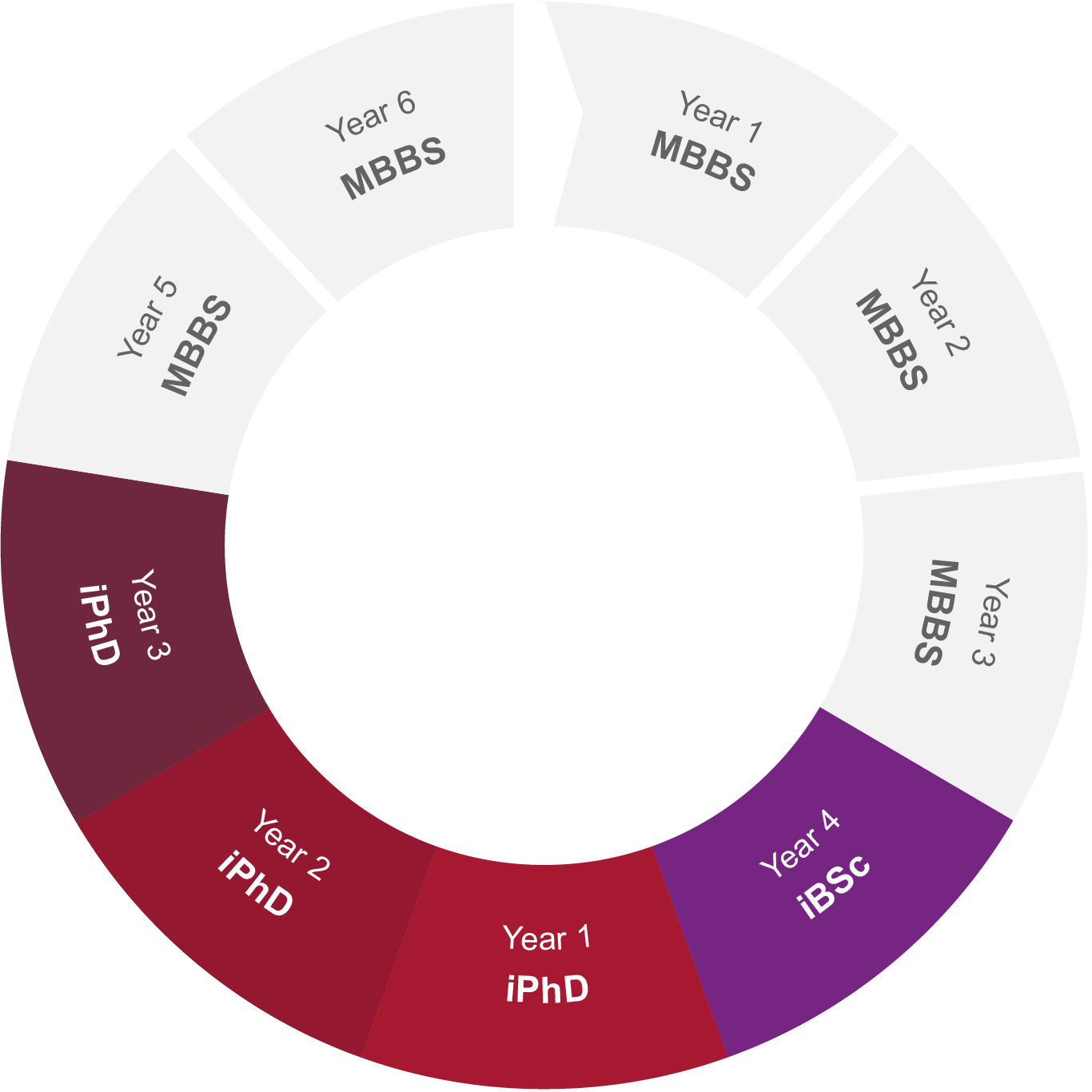
Support is available for 5 iPhD studentships for 2026. Studentships are typically 3 years in duration and provide:
You'll also have access to our Centre's training programme, available for all of our training programmes. See here for more information.
We welcome applications from students across all iBSc pathways, as long as they are committed to undertaking cancer research with a focus on convergence science.
1. Exploring project opportunities - October 2025
Imperial and ICR academics present summaries of their research projects. This is a chance to showcase the available research prospects for prospective students. The list of project summaries is available on this page every October.
Students review these project summaries and choose three projects they're interested in pursuing for their potential PhD. Once the review is done, students need to send their top three preferences using the Candidate Preference Submission Form - iPhD 2026 along with their CV and Equal Opportunities Form - iPhD 2026 to icr-imperial-convergence.centre@imperial.ac.uk. Project supervisors are then informed of students' interest in their projects, leading to meetings to discuss the opportunities.
3. Submit final project choices - 19th January 2026
After initial student-supervisor meetings, students rank the three preferred projects from 1 to 3 and share the rankings. The chosen supervisor is informed, and if they agree, they are invited to submit a full proposal.
4. Developing Proposals - January - April 2026
During this partnership, a comprehensive PhD project proposal is developed. The proposal is reviewed by the training committee who assess its scientific quality and its fit for a 3 year PhD.
If a student doesn't secure their top project choice, their second choice becomes available for consideration.
5. Full PhD Proposal Deadline - April 2026
6. Panel Interviews - May 2026
Next, shortlisted students, along with their supervisory team, participate in interviews. These interviews serve to assess a) the project's quality, suitability, and feasibility for a PhD, b) the support the supervisory team offers, and c) the student's motivations for pursuing a PhD.
7. PhD Commences - July 2026
Supervisors
Dr Claire Fletcher (Imperial, Department of Surgery and Cancer)
Proposed Outline
Prostate cancer (PC) affects 1-in-8 UK men. Despite major advances in precision oncology, metastatic PC (mPC) remains incurable. Standard-of-care chemotherapies confer only modest survival benefits and are associated with systemic toxicity and resistance development. There is an urgent need for efficacious, tumour-selective and resistance-resilient therapies.
We hypothesise that ‘programmable’ prostate-targeted nanogels can deliver potent miR therapeutics to PC tissues at high efficiency, providing proof-of-principle for future pre-clinical efficacy studies and clinical trials
Supervisors
Dr Prashant Srivastava (Imperial, National Heart & Lung Institute)
Proposed Outline
The project will develop predictive biomarkers for patient stratification to translate the novel targeted therapeutic, DTP3, into healthcare benefit in MM. DTP3 is a first-in-class GADD45β/MKK7 inhibitor in Phase-2 clinical development (EudraCT:2021-004028-13), having produced strong objective clinical responses without toxicities as monotherapy in heavily pretreated MM patients. DTP3 selectively blocks NF-κB-dependent survival in MM cells via a novel mode of action that targets the essential cancer-specific module, GADD45β/MKK7, downstream of NF-κB, rather than NF-κB itself, thus avoiding the dose-limiting toxicities of conventional IKK/NF-κB-targeting drugs [PMID:25314077;PMID:25314072]. As such, DTP3 selectively kills MM and other cancer cells ex-vivo and in-vivo with no toxicity to normal tissues and no adverse effects [PMID:25314077;PMID:31080744]. Having established DTP3’s clinical efficacy and tolerability in the unmet need of MM, we aim to develop precision biomarkers to improve MM treatment through better diagnostics by integrating multi-omics data (transcriptomics, genomics/methylomics, total/phospho-proteomics, scRNA-seq/scATAC-seq) from responders/non-responders in the current trial with similarly annotated public MM datasets [PMID:39160255;PMID:38942927].
Our previous work demonstrated that elevated GADD45B (and MKK7) expression predicts DTP3 response in primary MM cells ex-vivo [PMID:30255568]. However, genome-wide studies have shown that single-gene alterations fail to capture tumour heterogeneity or reliably predict drug-response in patients. To address this, we will apply deep-learning neural networks integrating multi-omics and single-cell data, alongside unsupervised methods (clustering, NMF), to deconvolute MM heterogeneity and identify biological feature/functional disease states linked to responsiveness/resistance to DTP3. Publicly available multi-omics MM datasets [PMID:39160255;PMID:38942927;PMID:37081258] will be used for training to map GADD45β/MKK7 activity to biological pathways and MM subtypes for biomarker discovery. Datasets will be harmonized using multi-modal AI frameworks to derive robust, interpretable, and portable results. Integrative multi-omics data from patients in DTP3’s trial will be used for clinical validation. In addition to resolving phenotypic hallmarks of DTP3-sensitive myelomas, the integrative analysis of tumours that relapse after an initial response to DTP3 will clarify mechanisms of acquired resistance and expose co-vulnerabilities for combination-therapy selection. We will prospectively use this information to develop clinical-grade diagnostics for therapy-response prognostication, to inform the individual treatment strategy and deliver an effective precision medicine that benefits patients with MM and other hard-to-treat, NF-κB-driven cancers.
Supervisors
Prof Oscar Ces (Imperial, Department of Chemistry)
Proposed Outline
Advanced prostate cancer (PCa) remains lethal, despite increases in overall survival afforded by the development of multiple new treatments including taxane chemotherapy, androgen receptor pathway inhibitors, PARP inhibitors and lutetium PSMA, due to eventual, inevitable treatment resistance. Thus, the most urgent unmet clinical need in inoperable, therapy resistant PCa is development of innovative treatment strategies with novel mechanisms of action that are effective and well-tolerated. Current therapies are associated with undesirable side-effects and systemic toxicities which decrease compliance and adversely affect patients’ quality of life. We have identified novel anti-apoptotic treatment strategies targeting the MCL1 alone, or in synergistic combinations, that drive cancer specific cell death in lethal PCa. They have toxicity profiles that make systemic delivery challenging. We propose to address this by loading drugs into “synthetic cells” that can respond specifically to the prostate tumour microenvironment to release their payload in a targeted, tumour cell-specific manner.
Supervisors
Dr Mitchell Chen (Imperial, Department of Surgery and Cancer)
Proposed Outline
Supervisors
Prof Dennis Wang (Imperial, National Heart & Lung Institute)
Proposed Outline
Supervisors
Dr Ehsan Ghorani (Imperial, Department of Surgery & Cancer)
Proposed Outline
For more information on this opportunity send an email to icr-imperial-convergence.centre@imperial.ac.uk.